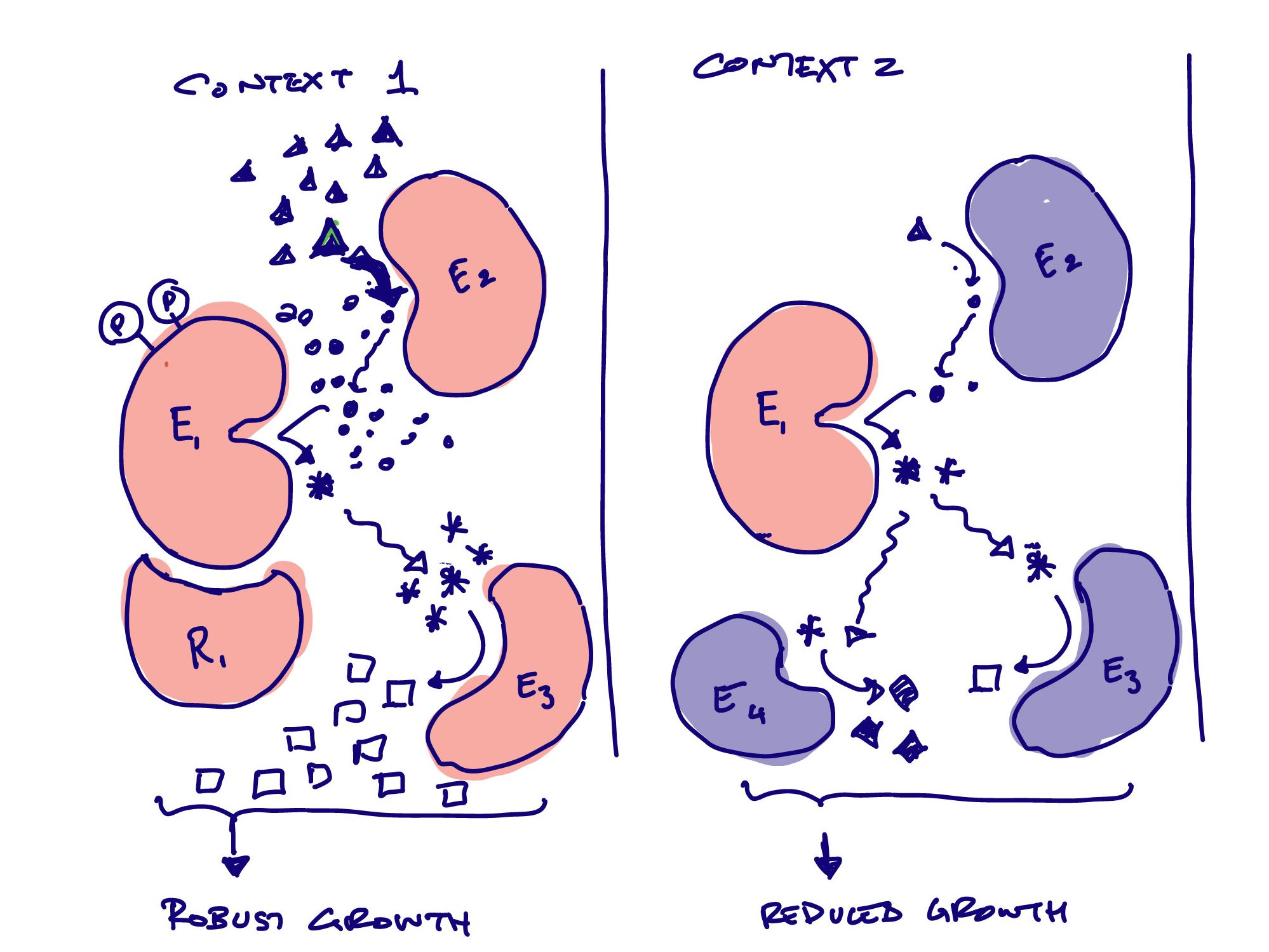
Modeling and Designing Proteins in Context.
Whether proteins assemble into a larger physical complexes, exchange intermediates in a metabolic pathway, or activate one another in a signaling cascade, protein-protein interactions form the basis for cells to grow, divide, and respond to the environment. In turn, cell context shapes the evolution and function of individual proteins.
Our goal is to design synthetic protein systems that function efficiently and appropriately when placed into an interaction-rich cellular context. To do this, we combine high-throughput functional assays with computational modeling and machine learning.